
Through metabarcoding, targeted isolation, and whole genome sequencing, we identified diverse Pseudomonads that appear to interact with Psa across kiwifruit germplasm.
Dec 30, 2025
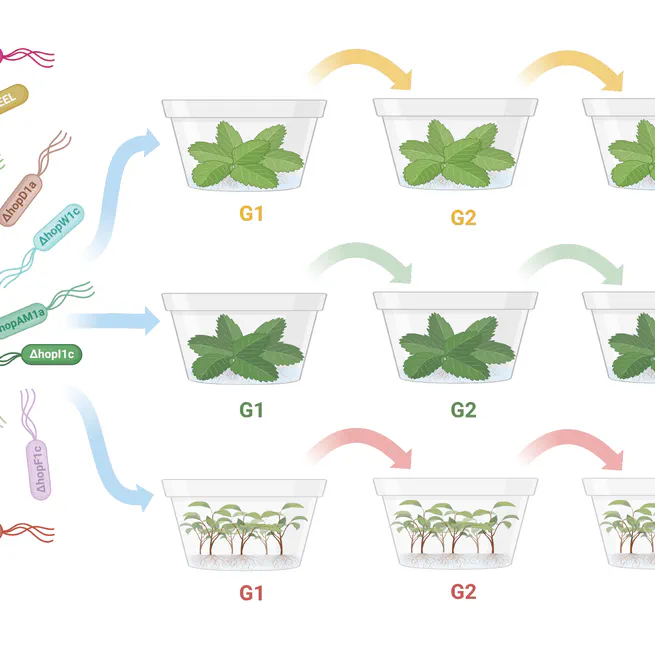
Through MELEE, a mutated effector-leveraging evolution experiment, we identified that the majority of Psa3's effectors are collectively required for virulence.
Nov 26, 2025
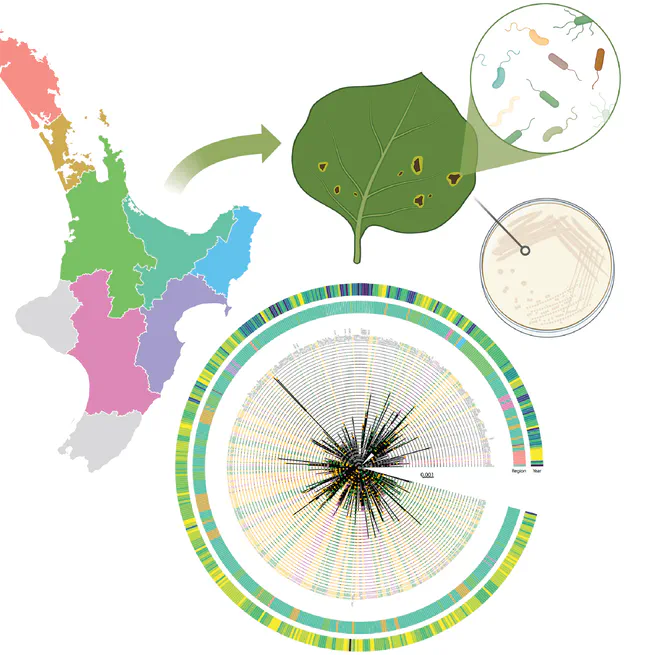
Genome biosurveillance of 500 Psa3 isolates over a 12-year period has revealed the emergence of hopF1c loss variants, mediated by the introduction of copper resistance elements into the population.
Feb 6, 2025

Through genome biosurveillance, effector screening, and transcriptomics, we characterised the immune response of resistant Actinidia melanandra to Psa3.
Oct 13, 2024
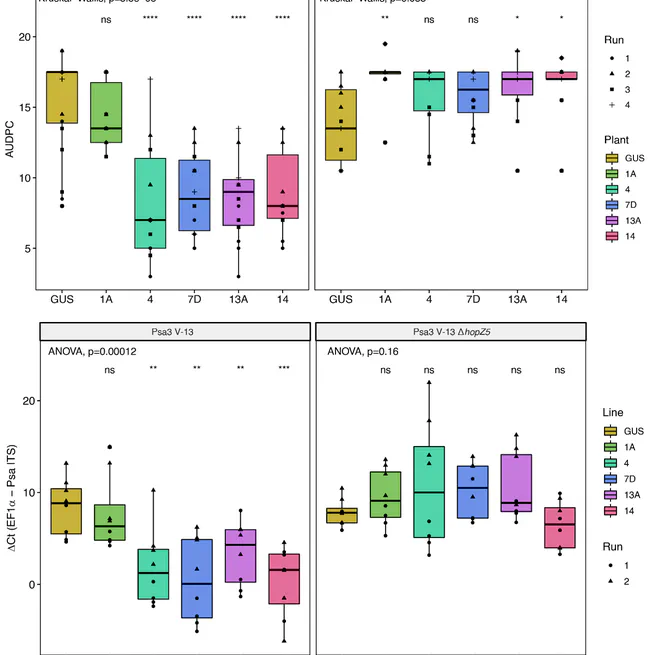
By transforming the PTR1 resistance gene from Nicotiana benthamiana into susceptible kiwifruit, we have demonstrated a potential strategy for engineering HopZ5-specific resistance against Psa3.
Jun 4, 2024
Effector knockout and knock-in assays in virulent Psa3 and avirulent Pfm identified a number of redundantly required effectors that converge on the RIN4 plant immunity hub.
Jan 1, 2023

Four avirulence effectors contribute to Psa3 recognition in resistant kiwiberry, identified through genome biosurveillance and effector knockout screening.
May 1, 2022
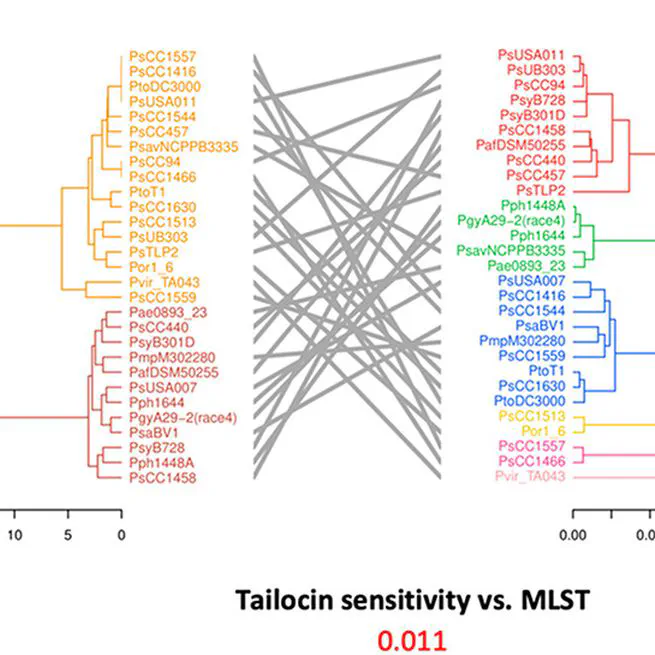
The TET operon determines the variability of LPS profiles in Psa, which is in turn correlated with tailocin susceptibility.
Dec 1, 2020