Genomic biosurveillance of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae biovar 3 reveals adaptation to selective pressures in New Zealand orchards
Feb 6, 2025· ,,,,,,,,,,,·
0 min read
,,,,,,,,,,,·
0 min read
Lauren Hemara
Stephen Hoyte
Saadiah Arshed
Magan Schipper
Peter Wood
Sergio Marshall
Mark Andersen
Haileigh Patterson
Joel Vanneste
Linda Peacock
Jay Jayaraman
Matthew Templeton
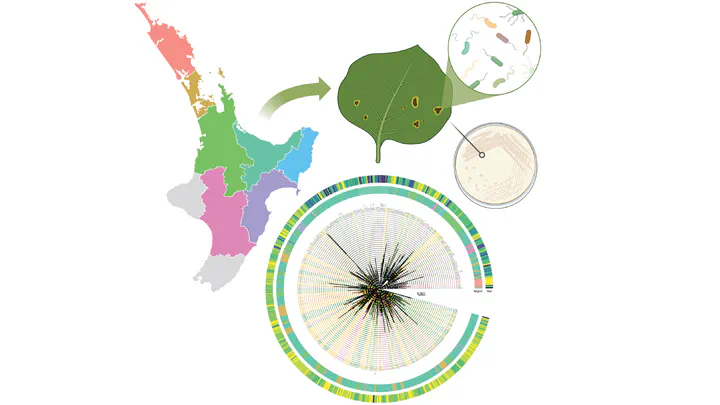
Abstract
In the late 2000s, a pandemic of Pseudomonas syringae pv. actinidiae biovar 3 (Psa3) devastated kiwifruit orchards growing susceptible yellow-fleshed cultivars. New Zealand’s kiwifruit industry has since recovered, following the deployment of the tolerant cultivar ‘Zesy002’. However, little is known about the extent to which the Psa population is evolving since its arrival. Over 500 Psa3 isolates from New Zealand kiwifruit orchards were sequenced between 2010 and 2022, from commercial monocultures and diverse germplasm collections. While effector loss was previously observed on Psa-resistant germplasm vines, effector loss appears to be rare in commercial orchards, where the dominant cultivars lack Psa resistance. However, a new Psa3 variant, which has lost the effector hopF1c, has arisen. The loss of hopF1c appears to have been mediated by the movement of integrative conjugative elements introducing copper resistance into this population. Following this variant’s identification, in planta pathogenicity and competitive fitness assays were performed to better understand the risk and likelihood of its spread. While hopF1c loss variants had similar in planta growth to wild-type Psa3, a lab-generated ∆hopF1c strain could outcompete wild-type on select hosts. Further surveillance was conducted in commercial orchards where these variants were originally isolated, with 6.6% of surveyed isolates identified as hopF1c loss variants. These findings suggest that the spread of these variants is currently limited, and they are unlikely to cause more severe symptoms than the current population. Ongoing genome biosurveillance of New Zealand’s Psa3 population is recommended to enable early detection and management of variants of interest.
Type
Publication
Molecular Plant Pathology
Genome Biosurveillance
Effector Recognition
Type III Effectors
Pseudomonas Syringae Pv. Actinidiae
Kiwifruit
Authors
Postdoctoral Fellow
Lauren Hemara is a Postdoctoral Fellow at the University of Toronto Scarborough.